This function creates a matrix where columns 1 and 2 correspond to tip labels and column 3 gives the depth of the MRCA of that pair of tips.
It is strongly based on treeVec and is used by relatedTreeDist and treeConcordance where tip labels belong to "categories".
tipsMRCAdepths(tree)Examples
tree <- rtree(10)
plot(tree)
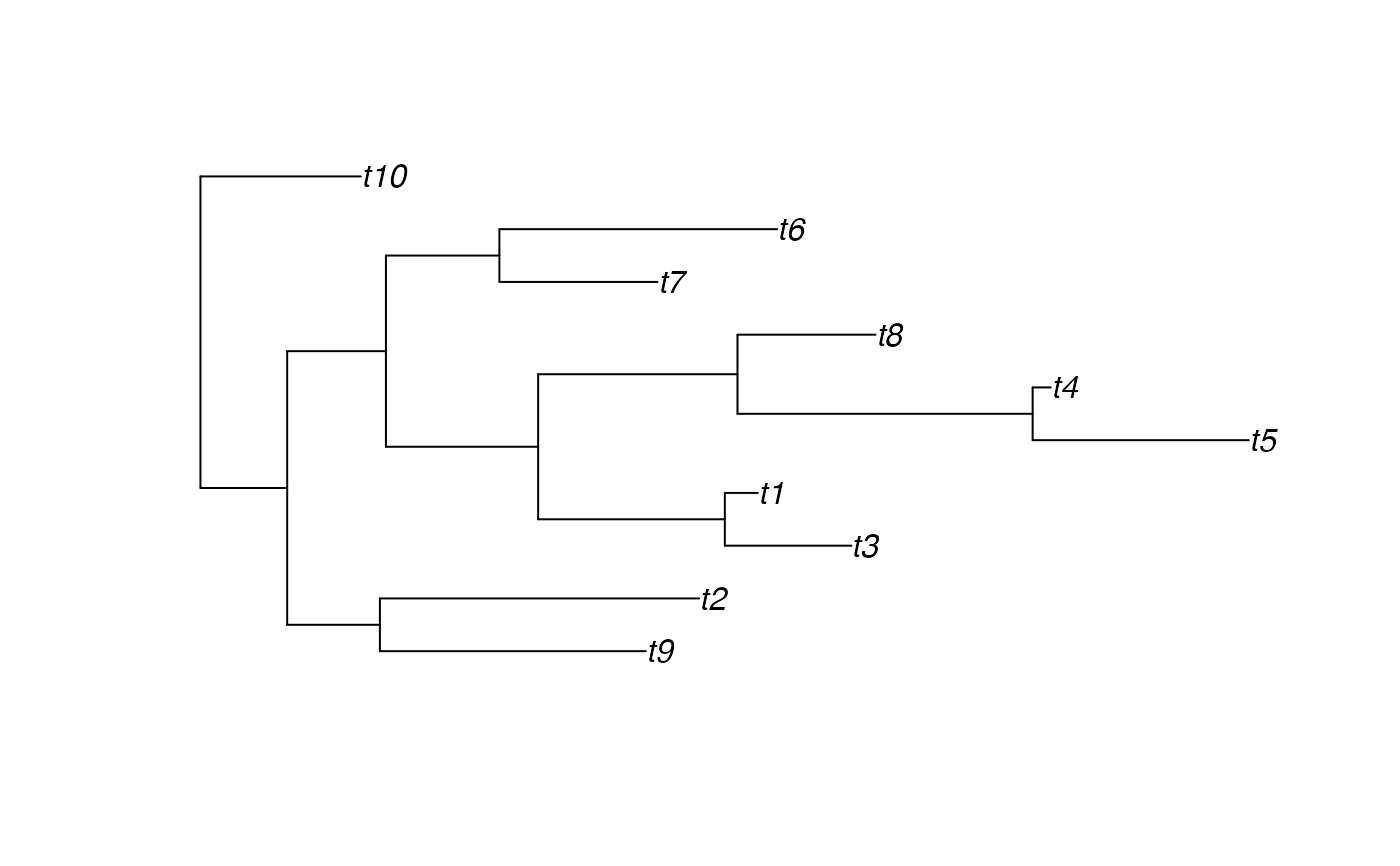 tipsMRCAdepths(tree)
#> tip1 tip2 rootdist
#> [1,] "t1" "t10" "1"
#> [2,] "t1" "t2" "0"
#> [3,] "t1" "t3" "0"
#> [4,] "t1" "t4" "1"
#> [5,] "t1" "t5" "1"
#> [6,] "t1" "t6" "1"
#> [7,] "t1" "t7" "1"
#> [8,] "t1" "t8" "0"
#> [9,] "t1" "t9" "0"
#> [10,] "t10" "t2" "0"
#> [11,] "t10" "t3" "0"
#> [12,] "t10" "t4" "2"
#> [13,] "t10" "t5" "2"
#> [14,] "t10" "t6" "2"
#> [15,] "t10" "t7" "2"
#> [16,] "t10" "t8" "0"
#> [17,] "t10" "t9" "0"
#> [18,] "t2" "t3" "2"
#> [19,] "t2" "t4" "0"
#> [20,] "t2" "t5" "0"
#> [21,] "t2" "t6" "0"
#> [22,] "t2" "t7" "0"
#> [23,] "t2" "t8" "2"
#> [24,] "t2" "t9" "1"
#> [25,] "t3" "t4" "0"
#> [26,] "t3" "t5" "0"
#> [27,] "t3" "t6" "0"
#> [28,] "t3" "t7" "0"
#> [29,] "t3" "t8" "3"
#> [30,] "t3" "t9" "1"
#> [31,] "t4" "t5" "3"
#> [32,] "t4" "t6" "3"
#> [33,] "t4" "t7" "3"
#> [34,] "t4" "t8" "0"
#> [35,] "t4" "t9" "0"
#> [36,] "t5" "t6" "5"
#> [37,] "t5" "t7" "4"
#> [38,] "t5" "t8" "0"
#> [39,] "t5" "t9" "0"
#> [40,] "t6" "t7" "4"
#> [41,] "t6" "t8" "0"
#> [42,] "t6" "t9" "0"
#> [43,] "t7" "t8" "0"
#> [44,] "t7" "t9" "0"
#> [45,] "t8" "t9" "1"
tipsMRCAdepths(tree)
#> tip1 tip2 rootdist
#> [1,] "t1" "t10" "1"
#> [2,] "t1" "t2" "0"
#> [3,] "t1" "t3" "0"
#> [4,] "t1" "t4" "1"
#> [5,] "t1" "t5" "1"
#> [6,] "t1" "t6" "1"
#> [7,] "t1" "t7" "1"
#> [8,] "t1" "t8" "0"
#> [9,] "t1" "t9" "0"
#> [10,] "t10" "t2" "0"
#> [11,] "t10" "t3" "0"
#> [12,] "t10" "t4" "2"
#> [13,] "t10" "t5" "2"
#> [14,] "t10" "t6" "2"
#> [15,] "t10" "t7" "2"
#> [16,] "t10" "t8" "0"
#> [17,] "t10" "t9" "0"
#> [18,] "t2" "t3" "2"
#> [19,] "t2" "t4" "0"
#> [20,] "t2" "t5" "0"
#> [21,] "t2" "t6" "0"
#> [22,] "t2" "t7" "0"
#> [23,] "t2" "t8" "2"
#> [24,] "t2" "t9" "1"
#> [25,] "t3" "t4" "0"
#> [26,] "t3" "t5" "0"
#> [27,] "t3" "t6" "0"
#> [28,] "t3" "t7" "0"
#> [29,] "t3" "t8" "3"
#> [30,] "t3" "t9" "1"
#> [31,] "t4" "t5" "3"
#> [32,] "t4" "t6" "3"
#> [33,] "t4" "t7" "3"
#> [34,] "t4" "t8" "0"
#> [35,] "t4" "t9" "0"
#> [36,] "t5" "t6" "5"
#> [37,] "t5" "t7" "4"
#> [38,] "t5" "t8" "0"
#> [39,] "t5" "t9" "0"
#> [40,] "t6" "t7" "4"
#> [41,] "t6" "t8" "0"
#> [42,] "t6" "t9" "0"
#> [43,] "t7" "t8" "0"
#> [44,] "t7" "t9" "0"
#> [45,] "t8" "t9" "1"