Comparison of two trees using the Kendall Colijn metric
treeDist(
tree.a,
tree.b,
lambda = 0,
return.lambda.function = FALSE,
emphasise.tips = NULL,
emphasise.weight = 2
)Arguments
- tree.a
an object of the class
phylo- tree.b
an object of the class
phylo(with the same tip labels as tree.a)- lambda
a number in [0,1] which specifies the extent to which topology (default, with lambda=0) or branch lengths (lambda=1) are emphasised. This argument is ignored if
return.lambda.function=TRUE.- return.lambda.function
If true, a function that can be invoked with different lambda values is returned. This function returns the vector of metric values for the given lambda.
- emphasise.tips
an optional list of tips whose entries in the tree vectors should be emphasised. Defaults to
NULL.- emphasise.weight
applicable only if a list is supplied to
emphasise.tips, this value (default 2) is the number by which vector entries corresponding to those tips are emphasised.
Value
The distance between the two trees according to the metric for the given value of lambda, or a function that produces the distance given a value of lambda.
Examples
## generate random trees
tree.a <- rtree(6)
tree.b <- rtree(6)
treeDist(tree.a,tree.b) # lambda=0
#> [1] 4.358899
treeDist(tree.a,tree.b,1) # lambda=1
#> [1] 2.60433
dist.func <- treeDist(tree.a,tree.b,return.lambda.function=TRUE) # distance as a function of lambda
dist.func(0) # evaluate at lambda=0. Equivalent to treeDist(tree.a,tree.b).
#> [1] 4.358899
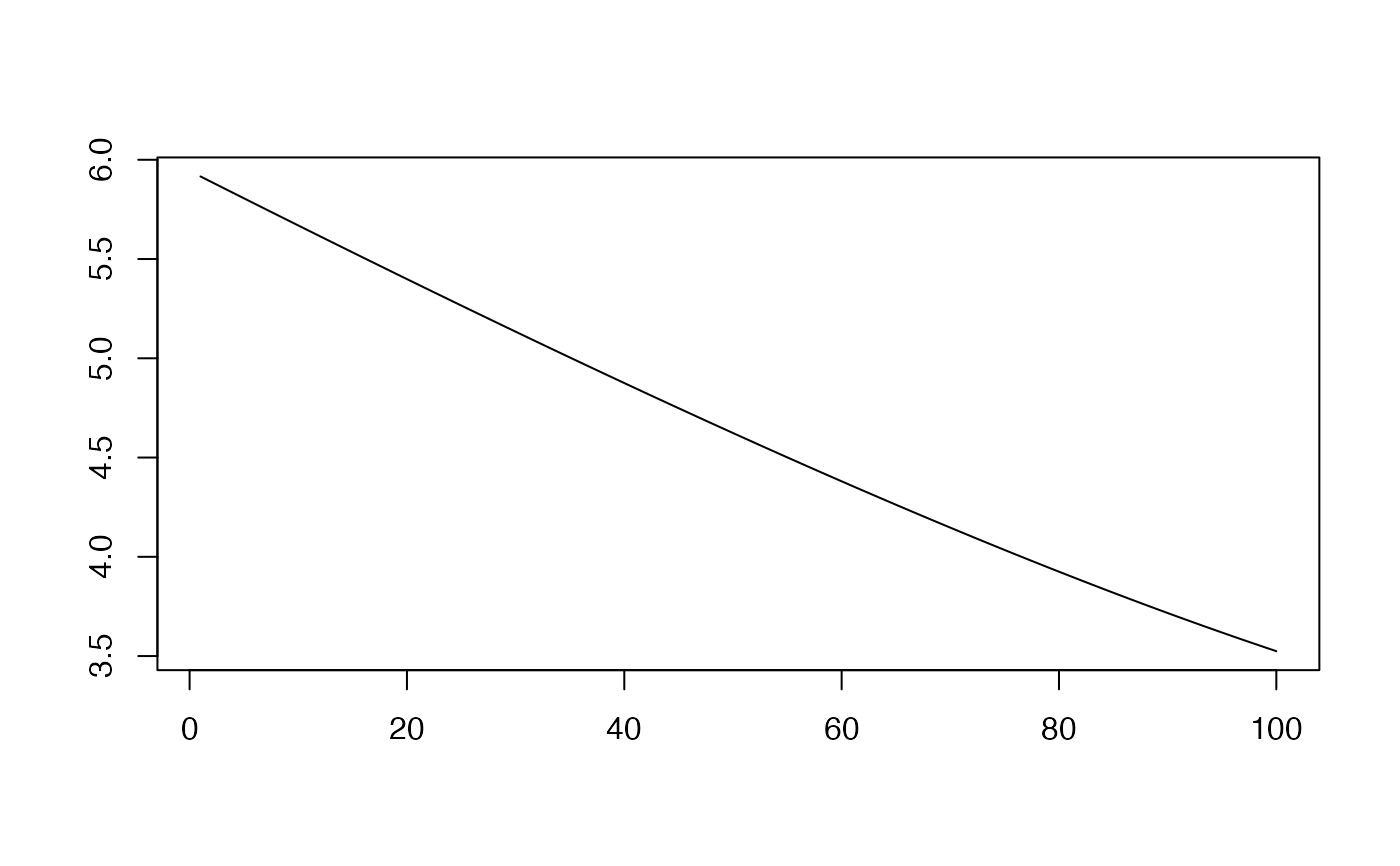
## We can see how the distance changes when moving from focusing on topology to length:
plot(sapply(seq(0,1,length.out=100), function(x) dist.func(x)), type="l",ylab="",xlab="")
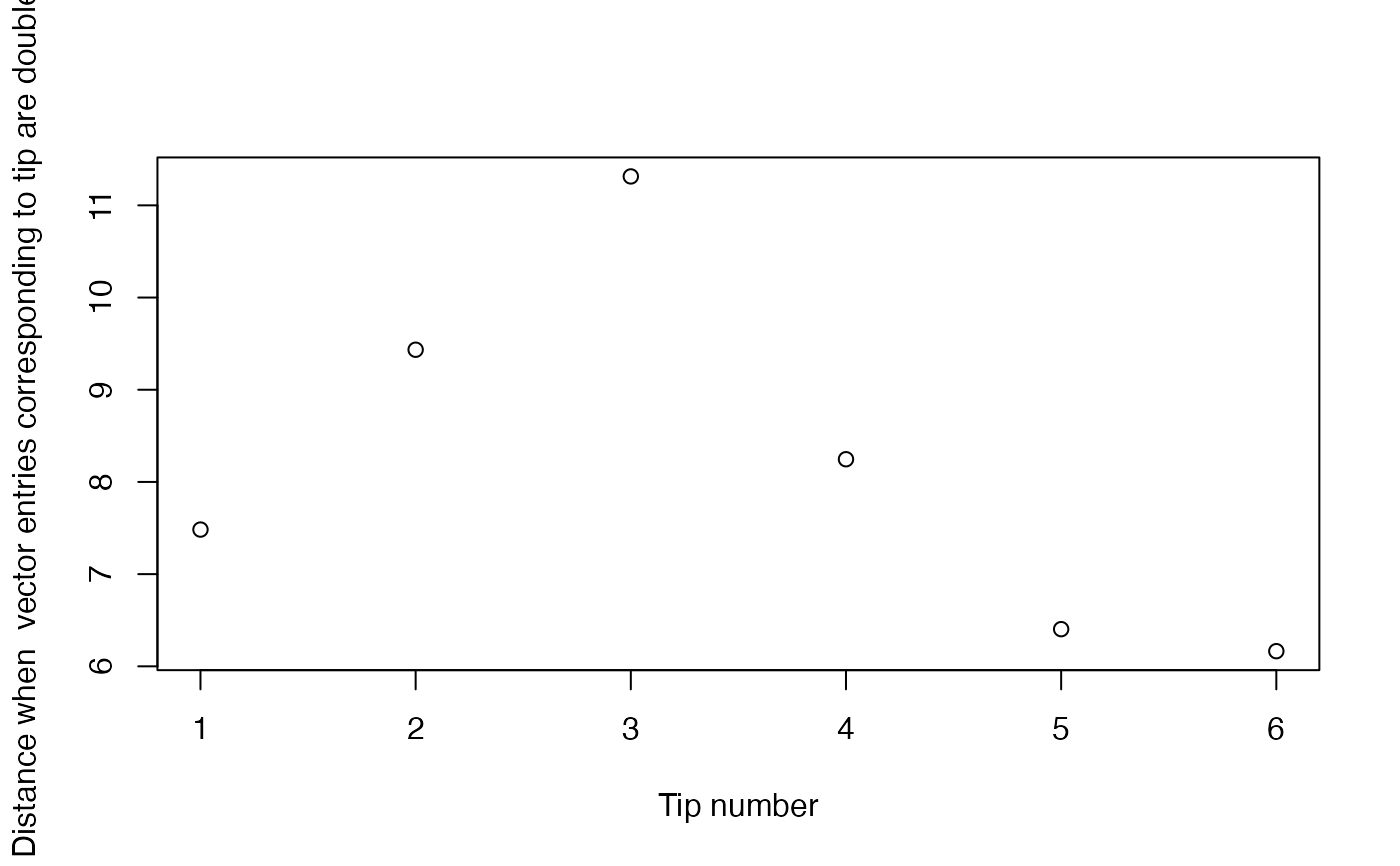
 ## The distance may also change if we emphasise the position of certain tips:
plot(sapply(tree.a$tip.label, function(x) treeDist(tree.a,tree.b,emphasise.tips=x)),
xlab="Tip number",ylab="Distance when vector entries corresponding to tip are doubled")
## The distance may also change if we emphasise the position of certain tips:
plot(sapply(tree.a$tip.label, function(x) treeDist(tree.a,tree.b,emphasise.tips=x)),
xlab="Tip number",ylab="Distance when vector entries corresponding to tip are doubled")