Reduce a tree with many tips into a tree with a single tip per category. Where a category's tips form a monophyletic clade, the clade is replaced by a single tip labelled by that category. Where a category's tips are paraphyletic, the largest clade for that category is treated as above, and all other tips pruned.
makeCollapsedTree(tree, df, warnings = TRUE)Arguments
Value
A tree (class phylo) whose tip labels are exactly the set of unique categories from df.
See also
Examples
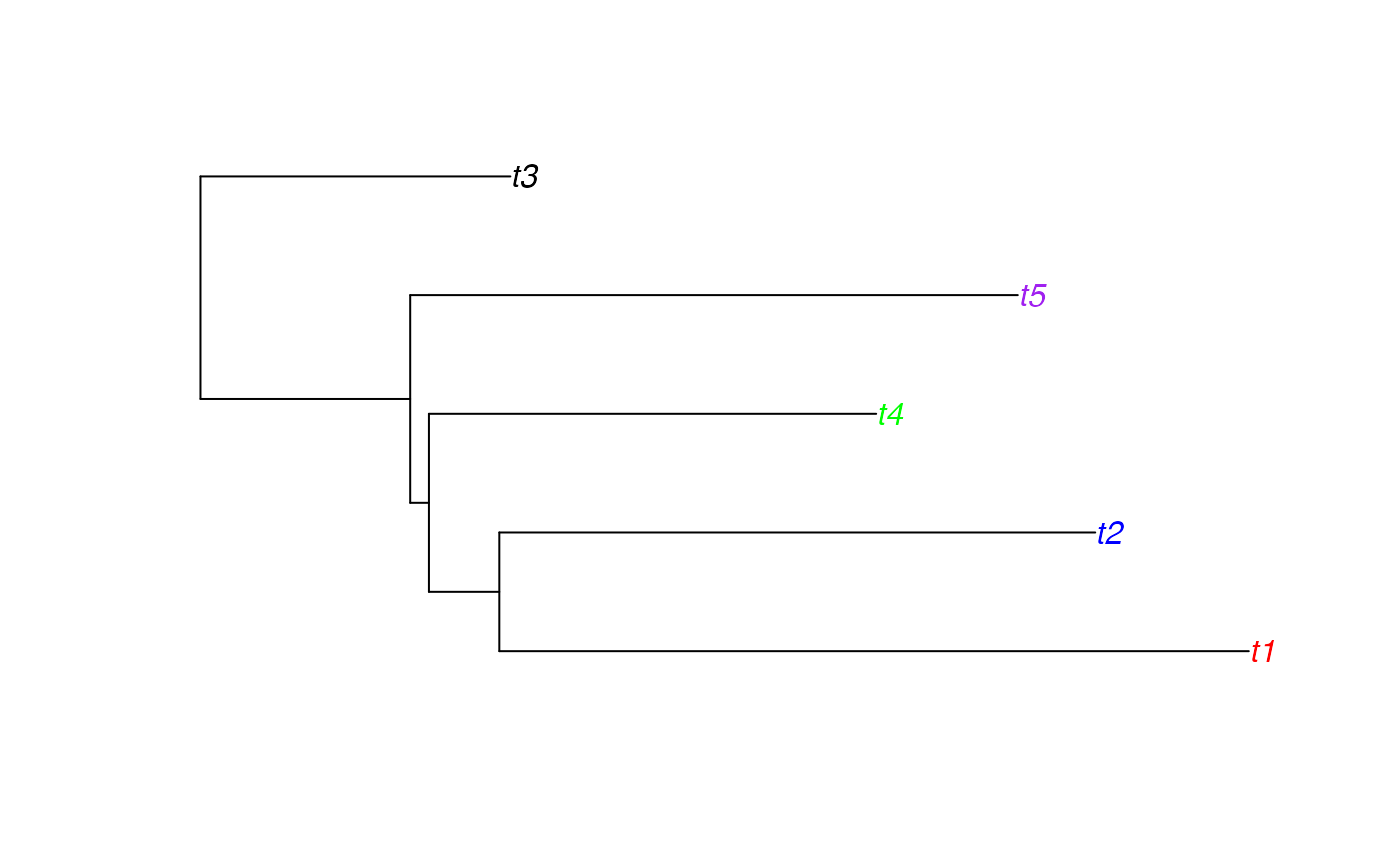
# simulate a tree which is monophyletic per category
tree <- simulateIndTree(rtree(5), permuteTips=FALSE)
df <- cbind(sort(rep(rtree(5)$tip.label,5)),sort(tree$tip.label))
palette <- c("red","blue","black","green","purple")#'
tipCols <- palette[as.factor(sapply(tree$tip.label, function(l) df[which(df[,2]==l),1]))]
plot(tree, tip.color=tipCols)
 collapsedTree <- makeCollapsedTree(tree,df)
plot(collapsedTree, tip.color=palette[as.factor(collapsedTree$tip.label)])
collapsedTree <- makeCollapsedTree(tree,df)
plot(collapsedTree, tip.color=palette[as.factor(collapsedTree$tip.label)])
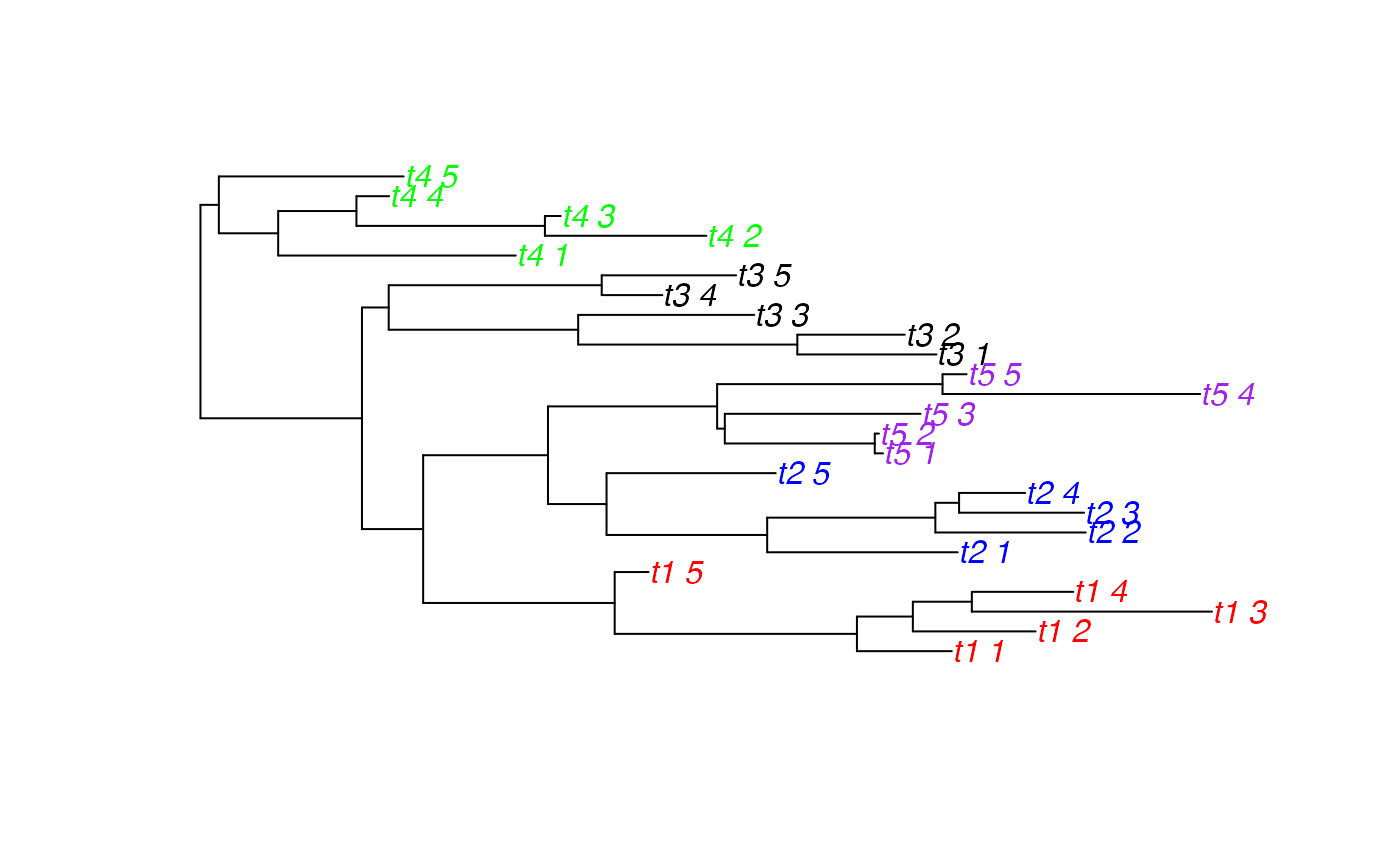
 # simulate a tree which is paraphyletic per category
tree <- simulateIndTree(rtree(5), tipPercent=20)
tipCols <- palette[as.factor(sapply(tree$tip.label, function(l) df[which(df[,2]==l),1]))]
plot(tree, tip.color=tipCols)
# simulate a tree which is paraphyletic per category
tree <- simulateIndTree(rtree(5), tipPercent=20)
tipCols <- palette[as.factor(sapply(tree$tip.label, function(l) df[which(df[,2]==l),1]))]
plot(tree, tip.color=tipCols)
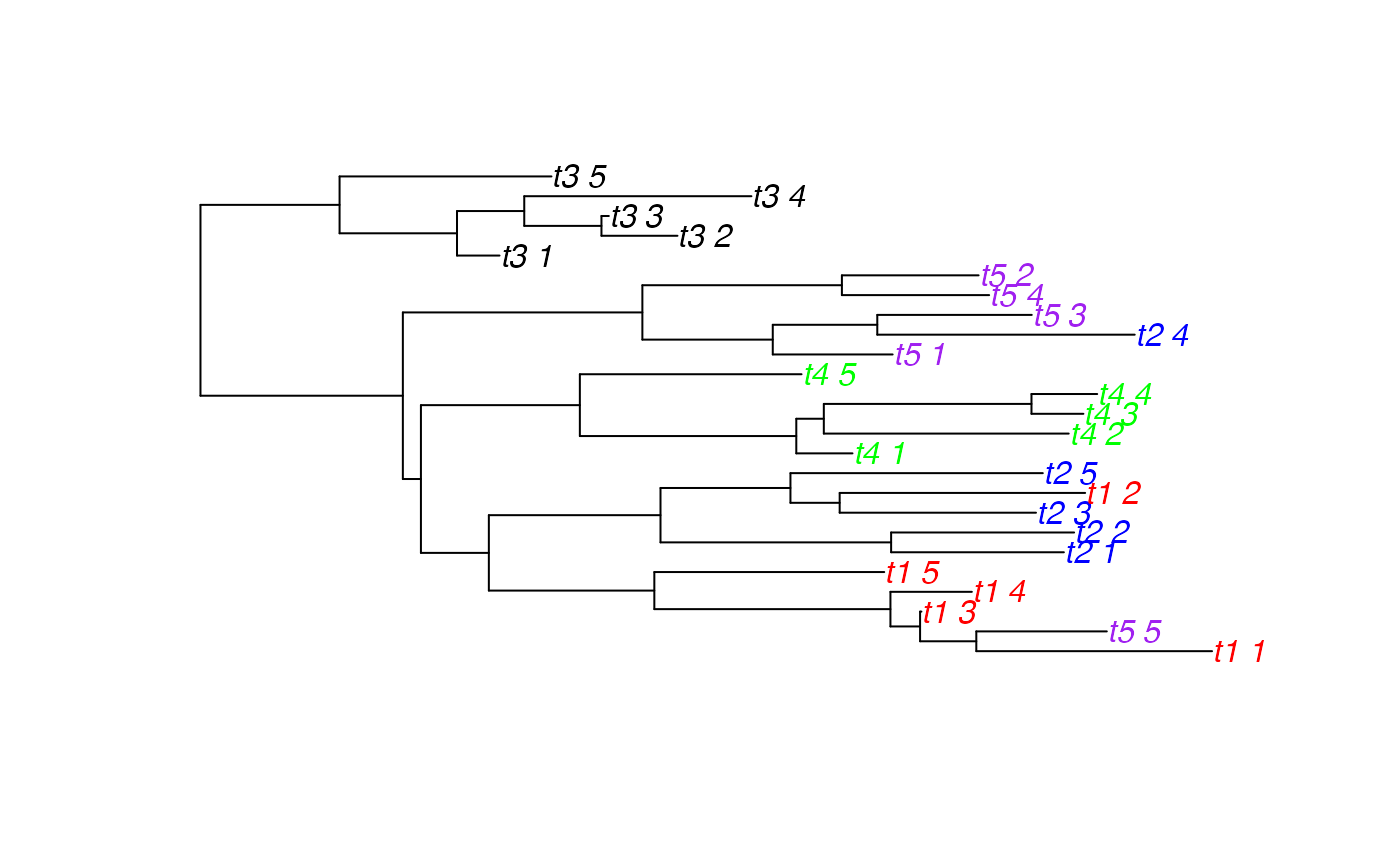 collapsedTree <- makeCollapsedTree(tree,df)
#> [1] "Note: the tree was not monophyletic per category"
plot(collapsedTree, tip.color=palette[as.factor(collapsedTree$tip.label)])
collapsedTree <- makeCollapsedTree(tree,df)
#> [1] "Note: the tree was not monophyletic per category"
plot(collapsedTree, tip.color=palette[as.factor(collapsedTree$tip.label)])