This function calculates the concordance between a category tree and an individuals tree
treeConcordance(catTree, indTree, df)Arguments
See also
Examples
# create an example category tree
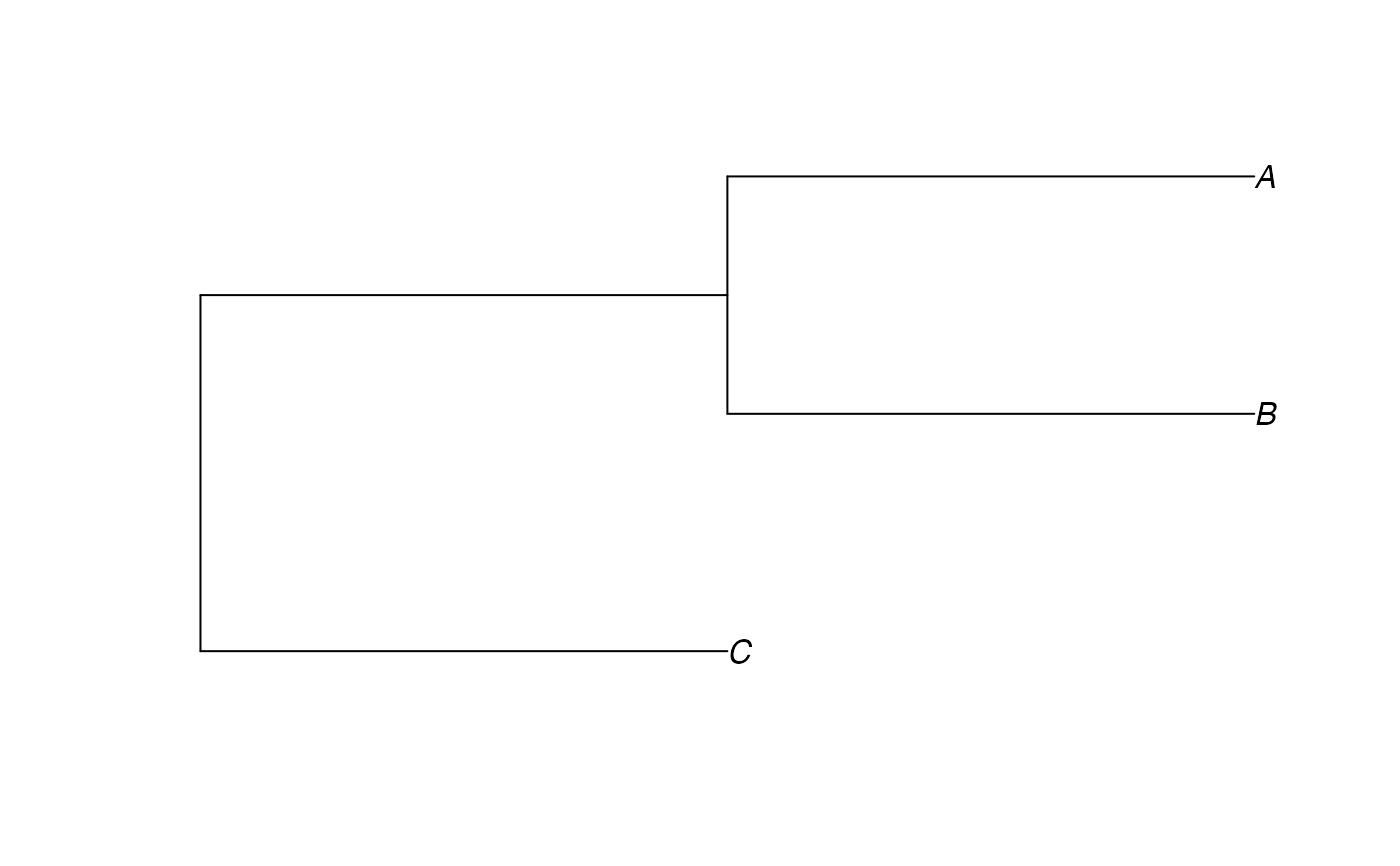
catTree <- read.tree(text="(C:1,(B:1,A:1):1);")
plot(catTree)
 # make individuals tree with complete concordance:
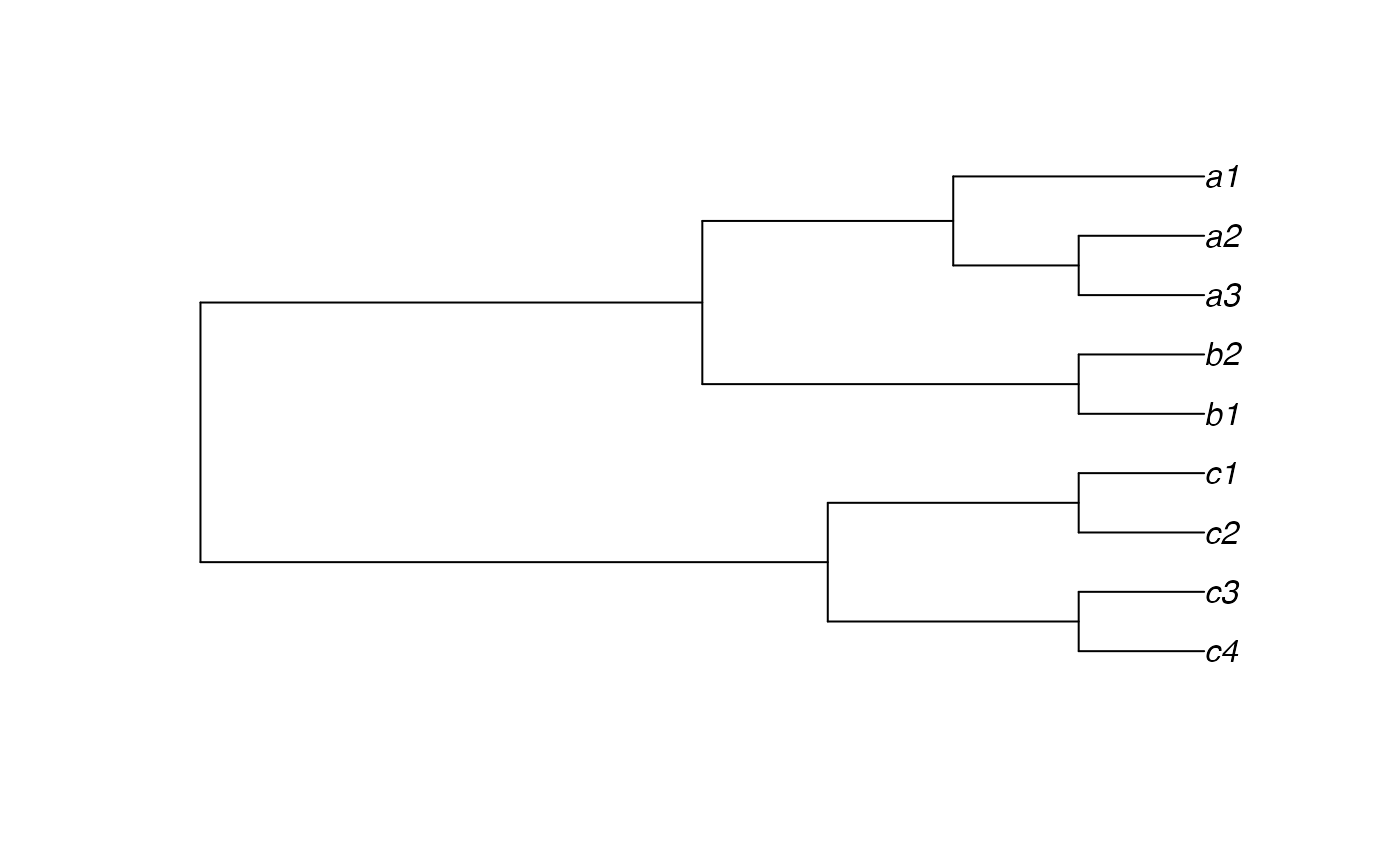
indTree1 <- read.tree(text="(((c4,c3),(c2,c1)),((b1,b2),((a3,a2),a1)));")
plot(indTree1)
# make individuals tree with complete concordance:
indTree1 <- read.tree(text="(((c4,c3),(c2,c1)),((b1,b2),((a3,a2),a1)));")
plot(indTree1)
 # create data frame linking categories with individuals
df <- cbind(c(rep("A",3),rep("B",2),rep("C",4)),sort(indTree1$tip.label))
treeConcordance(catTree,indTree1,df)
#> [1] 1
# make a less concordant tree:
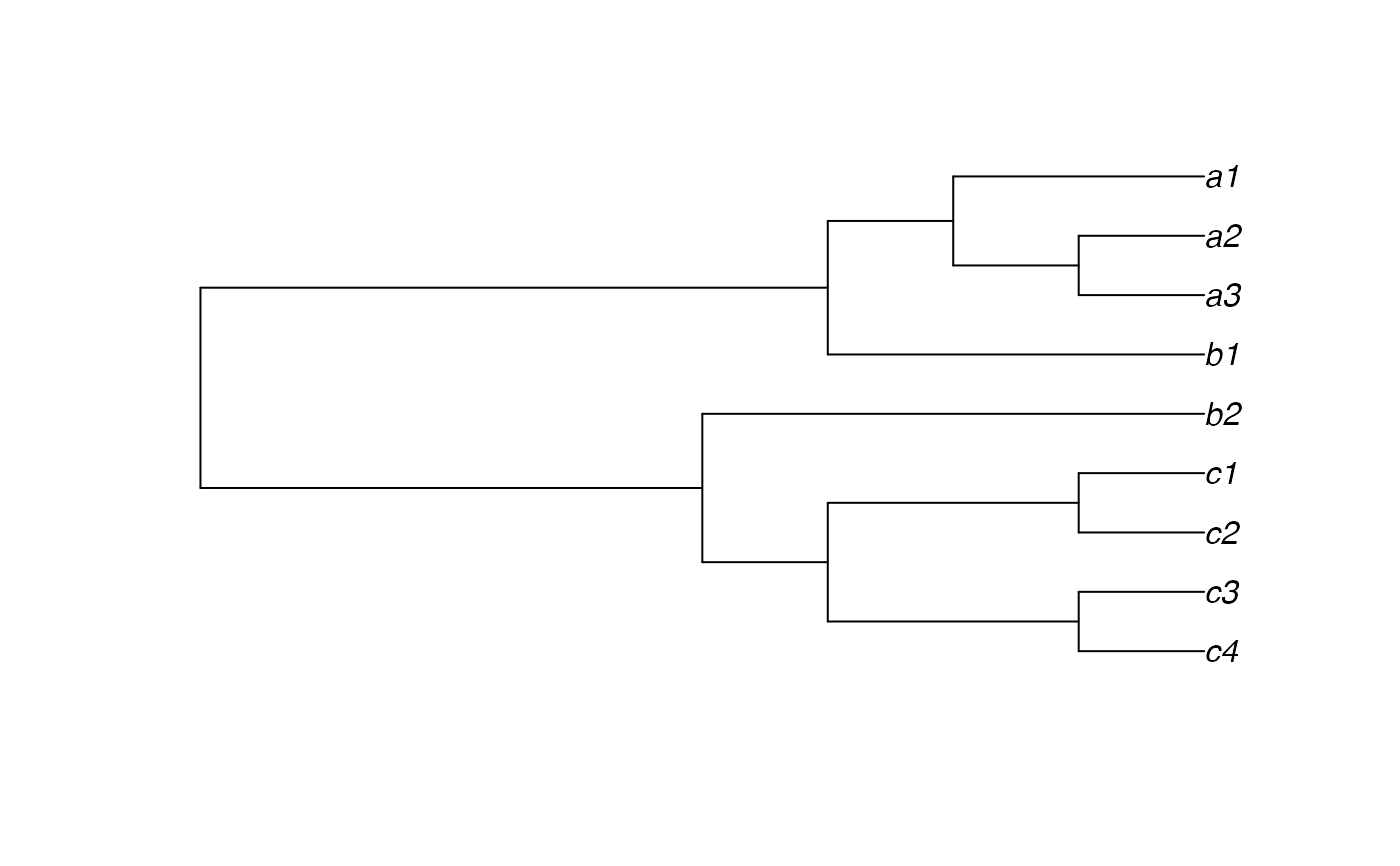
indTree2 <- read.tree(text="((((c4,c3),(c2,c1)),b2),(b1,((a3,a2),a1)));")
plot(indTree2)
# create data frame linking categories with individuals
df <- cbind(c(rep("A",3),rep("B",2),rep("C",4)),sort(indTree1$tip.label))
treeConcordance(catTree,indTree1,df)
#> [1] 1
# make a less concordant tree:
indTree2 <- read.tree(text="((((c4,c3),(c2,c1)),b2),(b1,((a3,a2),a1)));")
plot(indTree2)
 treeConcordance(catTree,indTree2,df)
#> [1] 0.7307692
# simulate larger example:
catTree <- rtree(10)
indTree3 <- simulateIndTree(catTree, tipPercent=10)
df <- cbind(sort(rep(catTree$tip.label,5)),sort(indTree3$tip.label))
plot(indTree3)
treeConcordance(catTree,indTree2,df)
#> [1] 0.7307692
# simulate larger example:
catTree <- rtree(10)
indTree3 <- simulateIndTree(catTree, tipPercent=10)
df <- cbind(sort(rep(catTree$tip.label,5)),sort(indTree3$tip.label))
plot(indTree3)
 treeConcordance(catTree,indTree3,df)
#> [1] 0.9075556
treeConcordance(catTree,indTree3,df)
#> [1] 0.9075556